WELCOME TO CilioGenics DATABASE
OR
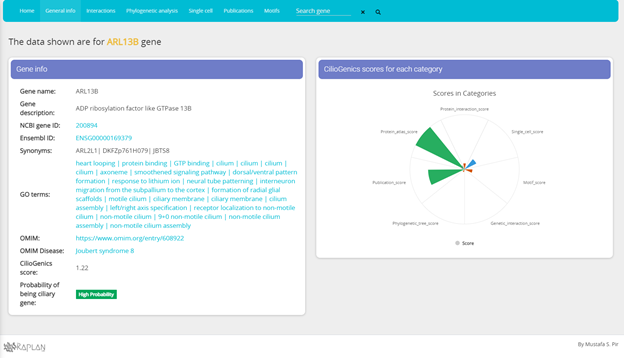
Gene info
CilioGenics scores for each category
* Integrated normalized CilioGenics scores.
* Motif score represents cilia associated TF (GLIS3, JAZF1, RFX2, RFX3, MYB and FOXJ1) interactions
* Phylogeny score is based on the cluster the searched gene belongs to.
* Protein and genetic interaction scores are based on the rate of interactions with ciliary proteins/genes by all interactions
Protein interactions
Network plot for protein interactions
Protein interactions
Genetic interactions
Network plot for genetic interactions
Genetic interactions
List of publications
Motifs
The table below shows the motifs that appears in the promoter (1000bp upstream) region of the gene of interest.
CilioGenics score table
This is an extensive list of all human genes included in the database. The column headers provide the description of the data, giving a score for each gene. Users can type a gene name and sort columns.
* Table shows weighted scores for all categories. Weights are calculated based on each section's capability to find ciliary genes.
Click the genes to easily switch to its gene search page.
Explore a compilation of 52 papers that presents the list of ciliary and putative ciliary genes.
List of publications
To explore the data sets, choose the article of interest from the table below.
* To open the paper on Pubmed instead, click title of the paper.
Motifs
Explore the motifs and the genes with these motifs in their promoter.
There are two usage of CilioGenics:
· Gene search: You can search a human gene by HGNC gene name, entrez gene id, synonym and Ensembl gene id. Genes in following organisms which have homolog in human can also be searched by gene name and gene id:
o Rattus norvegicus
o Mus musculus
o Drosophila melanogaster
o Danio rerio
o Caenorhabditis elegans
o Saccharomyces cerevisiae
· Explore data: Whole data can also be explored without having to specify a gene.
* Main gene search page
After searching the gene, further data can be explored about that gene.
* Various data can be selected to explore data
In general info page, you can find various information about the gene, including CilioGenics score for each different categories.
You can then select different tabs to examine the data about that gene further.
![]()
* Different tabs can be selected, each one providing information about searched gene.
Explore Data page:
Unlike the Gene Search page, Explore Data page provide more general information about the data.
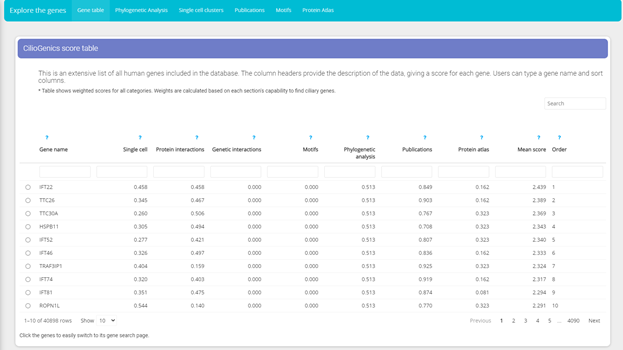
* CilioGenics score table shows scores from all categories.
Just like “Gene search” page, Explore data page also have multiple tabs to select. Tabs are mostly the same with “Gene search” page. However, in this page, you can explore data without having to specify any gene, giving the advantage of discovering new genes for a particular feature (like discovering genes having RFX1 motif in their promoters).
About CilioGenics
A Cilia Database
CilioGenics is an integrated and open source, community friendly database for ciliary genes. The key goal is to combine a variety of cilia-related data, including single-cell RNA-seq, comparative genomics, aggregation of gene list from cilia-related publications, protein atlas, proteomics, and cilia-specific motifs. Gene search is quick and user friendly, and it connects to a page devoted to the specific gene, which displays a variety of cilia-related information about that gene.
CilioGenics also provides a number of resources to:
explore the evolutionary dynamics ( the comparative genomics) of each gene in the interactive heatmap
explore the expression patterns of each gene (whole organisms in C. elegans, and lung tissue in human)
view proteomics relationship between ciliary proteins and other proteins
search for the name of gene in each cilia related publications
Update Policy of CilioGenics
Cilia are a complex organelle that possibly house hundreds of various proteins, and CilioGenics is a user-friendly database that aims to provide all existing knowledge about a gene. An automatic update is introduced into CilioGenics. For example, proteomics database BioGRID is automatically updated when a new update from BioGRID is released. However, since any effort to reveal the full constituents of cilia by one technique or a combination of two techniques is likely to fail to catch the entire ciliary genes and there will always new data about a gene, please help us to update the website if you have new data about a gene(s), as CilioGenics strives to incorporate new emerging data about each gene.
Make a Contribution to CilioGenics
We would be glad if you would contribute to expanding and improving the CilioGenics database. Here are a few ways you can help CilioGenics:
Submit a new paper presenting the ciliary gene list or single-cell RNA-seq paper.
Please submit a paper that is not currently in the CilioGenics collection, along with a brief description of the paper, such as a DOI for the paper and/or the integration table number. Please email us if you are willing to submit previously unpublished work.
Update existing data on the CilioGenics
Please consider submitting a feedback if you find any missing or incorrect information.
Cite
If you use CilioGenics in your publication, please cite:
Mustafa S Pir, Efe Begar, Ferhan Yenisert, Hasan C Demirci, Mustafa E Korkmaz, Asli Karaman, Sofia Tsiropoulou, Elif Nur Firat-Karalar, Oliver E Blacque, Sukru S Oner, Osman Doluca, Sebiha Cevik, Oktay I Kaplan, CilioGenics: an integrated method and database for predicting novel ciliary genes, Nucleic Acids Research, Volume 52, Issue 14, 12 August 2024, Pages 8127–8145, https://doi.org/10.1093/nar/gkae554
CilioGenics Version
CilioGenics is a dynamic and a constantly updated database, and if a major update is made, a new version of the database will be made available.
Cookie Policy
We use cookies to recognize your device or browser to adjust the content accordingly.
Contact
Please contact oktay.kaplan\@agu.edu.tr or mustafapir29\@gmail.com for any question or inquiries.
Source Code
For source code of the website: thekaplanlab/CilioGenics-website (github.com)